Thanks for choosing the Documents API to build NLP solutions into your app or website. Getting started with a new API can be challenging, so we have created a step-by-step guide that walks you through how to make your first API calls and more.
| Version | 1.0 |
| Endpoint | /-api/documents/v1 |
Authentication
The current API supports Basic Authentication. Note that the username and password are secured via an HTTPS connection.
Import and annotate text
One of the most common scenarios using tagtog is to import text to tagtog. The text will be automatically annotated if you are using any of the mechanisms to annotate text automatically (dictionaries, tagtog ML or your own ML). The API is the perfect way to automate document imports. To import annotated documents, go to the section: Import annotated documents.
Plain text POST
Import plain text.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
text |
"Hello, World!" | Plain text | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
ann.json |
The format of the output you want to be returned by the API. API output formats. |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
folder |
pool |
mySubFolder | Folder to store the document to. More information. You can refer to a folder by index, full path, or simple name. |
format |
Depends on the input type. Check the default formats. | formatted |
Force the format of the input. More info. |
distributeToMembers |
- |
John,Laura |
Parameter that overrides the default project task distribution settings. The format is a comma-separated list of the project user members to distribute to, and only those. Moreover, three special values exist: 1) This parameter is useful to fine-control which documents should be distributed to which members, depending on some criteria. For example, you could distribute documents to different members depending on the upload folder. |
filename |
text.txt | myPlainTextFile.txt | Force the document's filename with this argument, otherwise the default is used. Note that the filename must end with the extension .txt. Otherwise, this is appended to your given name. |
Examples: send plain text
By default, plain text imported to tagtog uses the verbatim input format. You should use this default mode when you want to keep the same formatting as your input text.
The example below imports plain text and retrieves the annotations identified (if any) in ann.json format.
curl -u yourUsername:yourPassword -X POST -d 'text="Hello, World!"' 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "ann.json"}
payload = {"text": "Hello, World!"}
response = requests.post(tagtogAPIUrl, params=params, auth=auth, data=payload)
print(response.text)
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json', {
method: 'POST',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword'),
'Accept': 'application/json',
'Content-Type': 'application/json',
},
body: JSON.stringify({"text": "Hello, World!"})
}).then(response => response.json()).then(json => {
console.log(json);
}).catch(function(error) {
console.log('Error: ', error);
});
Response, output=ann.json
{
"anncomplete":false,
"sources":[],
"entities":
[
{ "classId":"e_1","part":"s1p1","offsets":[{"start":251, "text":"natural killer"}],"confidence":{"state":"pre-added", "who":["ml:dpeker","prob":0.3287},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O14763","url":null},"recName":"Tumor necrosis factor receptor superfamily member 10B","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3287}}}},
{ "classId":"e_1","part":"s1p1","offsets":[{"start":267,"text":"NK"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3287},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O14763","url":null},"recName":"Tumor necrosis factor receptor superfamily member 10B","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3287}}}}
],
"metas":{},
"relations":[],
"annotatable":{"parts":["s1h1","s1p1"]}
}
Examples: send plain text and format it
Use the input format formatted to clean and format your input.
This example imports plain text in formatted format and returns the result of the operation (output format null).
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"project": "yourProjectName", "owner": "yourUsername", "format": "formatted", "output": "null"}
payload = {
"text": "The film stars Leonardo DiCaprio, Brad Pitt and Margot Robbie"
}
response = requests.post(tagtogAPIUrl, params=params, auth=auth, data=payload)
print(response.text)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "text.txt",
"names": ["text.txt"],
"rawInputSizeInBytes": 61,
"tagtogID": "aumzCn3f5E9zDs4yihXZAipZjLx0-text.txt",
"result": "created",
"parsedTextSizeInBytes": 61
}],
"warnings": []
}
URL POST GET
Import the content of a URL (HTML or other file) and annotate it.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
url |
https://en.wikipedia.org/wiki/Autonomous_cruise_control_system | URL to annotate | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
weburl |
The format of the output you want to be returned by the API. API output formats. |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
folder |
pool |
mySubFolder | Folder to store the document to. More information. You can refer to a folder by index, full path, or simple name. |
distributeToMembers |
- |
John,Laura |
Parameter that overrides the default project task distribution settings. The format is a comma-separated list of the project user members to distribute to, and only those. Moreover, three special values exist: 1) This parameter is useful to fine-control which documents should be distributed to which members, depending on some criteria. For example, you could distribute documents to different members depending on the upload folder. |
filename |
The original file name | Autonomous_cruise_control_system.html | Force the document's filename with this argument, otherwise the default is used. Note that the filename must end with the original extension. Otherwise, this is appended to your given name. |
Examples: import a web page
The example below imports a URL and as the output, it retrieves the web link for the annotated document. That link redirects to the annotated document at the tagtog web app. You can use other output formats.
curl -u yourUsername:yourPassword -X POST 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&url=https://en.wikipedia.org/wiki/Autonomous_cruise_control_system&output=weburl'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "weburl", "url": "https://en.wikipedia.org/wiki/Autonomous_cruise_control_system"}
response = requests.post(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&url=https://en.wikipedia.org/wiki/Autonomous_cruise_control_system&output=weburl', {
method: 'GET',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword')},
}).then(response => response.text()).then(text => {
console.log(text);
}).catch(function(error) {
console.log('Error: ', error);
});
Examples: import a file by URL
The example below imports a file given by a URL. The content will be represented by the default format associated to the filetype, in this case markdown. You can import other type of files as PDF or txt.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null", "url": "https://raw.githubusercontent.com/oxford-cs-deepnlp-2017/lectures/master/README.md"}
response = requests.post(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "README.md",
"filenames": ["README.md"],
"names": ["README.md"],
"rawInputSizeInBytes": 19680,
"docid": "aZkhd3qmP2BRoXhTOhUMjuxrz31i-README.md",
"tagtogID": "aZkhd3qmP2BRoXhTOhUMjuxrz31i-README.md",
"result": "created",
"parsedTextSizeInBytes": 19566
}],
"warnings": []
}
Files POST
Import a file and annotate it.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
files |
text.txt, text2.txt | List of files to annotate. Supported file types | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
ann.json |
The format of the output you want to be returned by the API. API output formats. |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
folder |
pool |
myFolder | Folder to store the document to. More information. You can refer to a folder by index, full path, or simple name. |
format |
verbatim |
Force how the format of the inputted text should be interpreted; more info. | |
distributeToMembers |
- |
John,Laura |
Parameter that overrides the default project task distribution settings. The format is a comma-separated list of the project user members to distribute to, and only those. Moreover, three special values exist: 1) This parameter is useful to fine-control which documents should be distributed to which members, depending on some criteria. For example, you could distribute documents to different members depending on the upload folder. |
filename |
The original file name | MyNewDoc.pdf | Force the document's filename with this argument, otherwise the default is used. Note that the filename must end with the original extension. Otherwise, this is appended to your given name. |
Examples: import a plain text file
This example imports a file and retrieves the annotations in ann.json.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "ann.json"}
#you can append more files to the list in case you want to upload multiple files
files = [("files", open('files/text.txt'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
var input = document.querySelector('input[type="file"]')
var data = new FormData()
data.append("files", input.files[0])
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json', {
method: 'POST',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword')},
body: data
}).then(response => response.text()).then(text => {
console.log(text);
}).catch(function(error) {
console.log('Error: ', error);
});
curl -u yourUsername:yourPassword -X POST -F 'files=@/files/document.txt' 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json'
Response, output=ann.json
{
"anncomplete":false,
"sources":[],
"entities":[
{"classId":"e_1","part":"s1p5","offsets":[{"start":187,"text":"apolipoprotein E"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p5","offsets":[{"start":205,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p6","offsets":[{"start":0,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p7","offsets":[{"start":0,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p8","offsets":[{"start":0,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p9","offsets":[{"start":0,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p11","offsets":[{"start":24,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p11","offsets":[{"start":108,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p11","offsets":[{"start":139,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p11","offsets":[{"start":223,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p12","offsets":[{"start":41,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p12","offsets":[{"start":146,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p12","offsets":[{"start":180,"text":"APOE"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P02649","url":null},"recName":"Apolipoprotein E","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.9293}}}},
{"classId":"e_1","part":"s1p15","offsets":[{"start":0,"text":"ABCA7"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IZY2","url":null},"recName":"ATP-binding cassette sub-family A member 7","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271}}}},
{"classId":"e_1","part":"s1p15","offsets":[{"start":25,"text":"ABCA7"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IZY2","url":null},"recName":"ATP-binding cassette sub-family A member 7","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271}}}},
{"classId":"e_1","part":"s1p16","offsets":[{"start":0,"text":"CLU"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6826},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P10909","url":null},"recName":"Clusterin","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6826}}}},
{"classId":"e_1","part":"s1p17","offsets":[{"start":0,"text":"CR1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6826},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"P17927","url":null},"recName":"Complement receptor type 1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6826}}}},
{"classId":"e_1","part":"s1p18","offsets":[{"start":0,"text":"PICALM"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8617},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13492","url":null},"recName":"Phosphatidylinositol-binding clathrin assembly protein","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8617}}}},
{"classId":"e_1","part":"s1p19","offsets":[{"start":0,"text":"PLD3"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IV08","url":null},"recName":"Phospholipase D3","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s1p19","offsets":[{"start":51,"text":"PLD3"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IV08","url":null},"recName":"Phospholipase D3","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s1p20","offsets":[{"start":0,"text":"TREM2"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q9NZC2","url":null},"recName":"Triggering receptor expressed on myeloid cells 2","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271}}}},
{"classId":"e_1","part":"s1p21","offsets":[{"start":0,"text":"SORL1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q92673","url":null},"recName":"Sortilin-related receptor","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271}}}},
{"classId":"e_1","part":"s1p21","offsets":[{"start":26,"text":"SORL1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q92673","url":null},"recName":"Sortilin-related receptor","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.8271}}}}
],
"metas":{},
"relations":[],
"annotatable":{"parts":["s1h1","s1p1","s1p2","s1p3","s1p4","s1p5","s1p6","s1p7","s1p8","s1p9","s1p10","s1p11","s1p12","s1p13","s1p14","s1p15","s1p16","s1p17","s1p18","s1p19","s1p20","s1p21"]}
}
Examples: import a PDF file
This example imports a PDF file and retrieves the annotations in ann.json. Please notice we open the PDF file in binary format. You can extend it easily to upload multiple files.
curl -u yourUsername:yourPassword -X POST -F 'files=@/files/document.pdf' 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "ann.json"}
#you can append more files to the list in case you want to upload multiple files
files = [("files", open("files/document.pdf", "rb"))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=ann.json
{
"annotatable": {
"parts": ["s1v1", "s2v1", "s3v1", "s4v1", "s5v1", "s6v1", "s7v1", "s8v1", "s9v1", "s10v1", "s11v1", "s12v1", "s13v1", "s14v1"]
},
"anncomplete": false,
"sources": [],
"metas": {},
"entities": [],
"relations": []
}
Examples: import a markdown file
This example imports a markdown file. You can also import a txt file and force the format to markdown.
Using Markdown you can also use tagtog blocks to build a customized annotation layout for your project! E.g. question answering datasets, chatbot training, tweets, etc.
curl -u yourUsername:yourPassword -X POST -F "files=@/files/readme.md" 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null"}
files = [("files", open('files/readme.md'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "README.md",
"filenames": ["README.md"],
"names": ["README.md"],
"rawInputSizeInBytes": 19680,
"docid": "aZkhd3qmP2BRoXhTOhUMjuxrz31i-README.md",
"tagtogID": "aZkhd3qmP2BRoXhTOhUMjuxrz31i-README.md",
"result": "created",
"parsedTextSizeInBytes": 19566
}],
"warnings": []
}
Examples: import a list of files
This example imports a list of plain text files (it can be any other supported file type or a combination) and retrieves the result of the operation.
curl -u yourUsername:yourPassword -X POST -F "files=@/files/item1.txt" -F "files=@/files/item2.txt" -F "files=@/files/item3.txt" 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null"}
files = [("files", open('files/item1.txt')), ("files", open('files/item2.txt')), ("files", open('files/item3.txt'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=null
{
"ok": 3,
"errors": 0,
"items": [{
"origid": "item1.txt",
"filenames": ["item1.txt"],
"names": ["item1.txt"],
"rawInputSizeInBytes": 128,
"docid": "aGMgsSYn0VJlSHWgGD4zwsIvOqDG-item1.txt",
"tagtogID": "aGMgsSYn0VJlSHWgGD4zwsIvOqDG-item1.txt",
"result": "created",
"parsedTextSizeInBytes": 128
}, {
"origid": "item2.txt",
"filenames": ["item2.txt"],
"names": ["item2.txt"],
"rawInputSizeInBytes": 53,
"docid": "aNkqrGOQX49FemNFJhx5GgPc9UAS-item2.txt",
"tagtogID": "aNkqrGOQX49FemNFJhx5GgPc9UAS-item2.txt",
"result": "created",
"parsedTextSizeInBytes": 53
}, {
"origid": "item3.txt",
"filenames": ["item3.txt"],
"names": ["item3.txt"],
"rawInputSizeInBytes": 41,
"docid": "azUkkxgJ7taVY7mzM71ciFKwp27i-item3.txt",
"tagtogID": "azUkkxgJ7taVY7mzM71ciFKwp27i-item3.txt",
"result": "created",
"parsedTextSizeInBytes": 39
}],
"warnings": []
}
PubMed Abstracts POST GET
Import one or more PubMed abstracts and annotate them.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
idType |
tagtogID |
PMID |
Type of Id. List of idTypes |
ids |
23596191, 29438695 | Comma-separated list of ids, all the same type. The response is limited to the last id imported. | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
ann.json |
The format of the output you want to be returned by the API. API output formats. |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
folder |
pool |
myFolder | Folder to store the document to. More information. You can refer to a folder by index, full path, or simple name. |
distributeToMembers |
- |
John,Laura |
Parameter that overrides the default project task distribution settings. The format is a comma-separated list of the project user members to distribute to, and only those. Moreover, three special values exist: 1) This parameter is useful to fine-control which documents should be distributed to which members, depending on some criteria. For example, you could distribute documents to different members depending on the upload folder. |
filename |
The original file name | myPaper.xml | Force the document's filename with this argument, otherwise the default is used. Note that the filename must end with the original extension. Otherwise, this is appended to your given name. |
Examples: import a list of PubMed articles by PMID
The example below imports a list of PMIDs and retrieves the annotations of the last document in ann.json format.
curl -u yourUsername:yourPassword -X POST 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&idType=PMID&ids=23596191,29438695&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'idType':'PMID', 'ids':['23596191','29438695'], "output": "ann.json"}
response = requests.post(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
fetch('https://www.tagtog.com/api/0.1/documents?project=yourProject&owner=yourUsername&idType=PMID&ids=23596191,29438695&output=ann.json', {
method: 'POST',
headers: { 'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword'),
'Accept': 'application/json',
'Content-Type': 'application/json',
},
}).then(response => response.json()).then(json => {
console.log(json);
}).catch(function(error) {
console.log('Error: ', error);
});
Response, output=ann.json
{
"anncomplete":false,
"sources":[{"name":"PMID","id":"23596191","url":"http://www.ncbi.nlm.nih.gov/pubmed/23596191"}],
"entities":
[
{"classId":"e_1","part":"s1h1","offsets":[{"start":60,"text":"RETICULATA-RELATED"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":315,"text":"RETICULATA-RELATED"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":335,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":444,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":561,"text":"PROTEIN"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3289},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IVL6","url":null},"recName":"Prolyl 3-hydroxylase 3","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3289}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1127,"text":"rer1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4836},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4836}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1265,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1303,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1391,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1587,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1591,"text":"proteins"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4073},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q15517","url":null},"recName":"Corneodesmosin","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4073}}}}
],
"metas":{},
"relations":[],
"annotatable":{"parts":["s1h1","s2h1","s2p1"]}
}
Import annotated documents POST
If you have annotated documents you want to import, you need to upload two files:
The text or document. This can be a regular file (e.g. txt, xml, pdf, plain.html, etc.), plain text, etc. Check the supported input types
The annotations. You pass this as an ann.json.
They must have the same name, except for the file extensions. For example: mydoc.pdf and mydoc.ann.json.
You can use the same API method you use to upload a single file to annotate: Files API POST.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
files |
text.txt, text.ann.json | You need to upload in the same request both: the text (supported input format) and the ann.json (annotations) files. | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
null |
|
format |
No default for pre-annotated documents, you should always set this parameter | default-plus-annjson |
Format of the pre-annotated document. List of supported pre-annotated formats: Pre-annotated formats |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
folder |
pool |
myFolder | Folder to store the document to. More information. You can refer to a folder by index, full path, or simple name. |
distributeToMembers |
- |
John,Laura |
Parameter that overrides the default project task distribution settings. The format is a comma-separated list of the project user members to distribute to, and only those. Moreover, three special values exist: 1) This parameter is useful to fine-control which documents should be distributed to which members, depending on some criteria. For example, you could distribute documents to different members depending on the upload folder. |
filename |
Name of the file imported | myPlainTextFile.txt | Force the document's filename with this argument, otherwise the default is used. Note that the filename must end with the original file extension. Otherwise, this is appended to your given name. |
Examples: import pre-annotated plain text file
This example shows how to upload a preannotated document (txt file + ann.json) to tagtog. The format used is default-plus-annjson to indicate we are importing pre-annotated content, the text content will be represented using the default format. In this case, the default format for plain text is verbatim. Make sure the ann.json is well formated according to the ann.json specification.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null", 'format': 'default-plus-annjson'}
files=[("files", open('files/text.txt')), ("files", open('files/text.ann.json'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
curl -u yourUsername:yourPassword -X POST -F "files=@/files/item1.txt" -F "files=@/files/item1.ann.json" 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&format=default-ann-json&output=ann.json'
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "text.txt",
"filenames": ["text.txt", "text.ann.json"],
"names": ["text.txt", "text.ann.json"],
"rawInputSizeInBytes": 1048102,
"docid": "aqXHSykmx2gmA9AJXW38OAl0DnTe-text.txt",
"tagtogID": "aqXHSykmx2gmA9AJXW38OAl0DnTe-text.txt",
"result": "created",
"parsedTextSizeInBytes": 81360
}],
"warnings": []
}
Examples: import pre-annotated raw plain text
This example shows how to upload a preannotated document (plain text + ann.json) to tagtog. The format used is default-plus-annjson to indicate we are importing pre-annotated content, the text content will be represented using the default format. In this case, the default format for plain text is verbatim. Make sure the ann.json is well formated according to the ann.json specification. In this example, we put directly in the code the plain text and the ann.json. It might be useful if you don't want to store this content on physical files.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null", 'format': 'default-plus-annjson'}
#you could easily point to an existing ann.json file or text file. e.g.: ("files", open('files/text.ann.json'))
files=[('hellotag.txt', 'Hello tag world'), ('hellotag.ann.json', '{"annotatable": {"parts": ["s1v1"]},"anncomplete": false,"sources": [],"metas": {},"entities": [{"classId": "e_1","part": "s1v1","offsets": [{"start": 6,"text": "tag"}],"confidence": {"state": "pre-added","who": ["user:yourUsername"],"prob": 1},"fields": {},"normalizations": {}}],"relations": []}')]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "hellotag.txt",
"filenames": ["hellotag.ann.json", "hellotag.txt"],
"names": ["hellotag.ann.json", "hellotag.txt"],
"rawInputSizeInBytes": 307,
"docid": "awq0S.5DQRW3Cjpv4u1tJyXl.L3m-hellotag.txt",
"tagtogID": "awq0S.5DQRW3Cjpv4u1tJyXl.L3m-hellotag.txt",
"result": "created",
"parsedTextSizeInBytes": 15
}],
"warnings": []
}
Examples: import pre-annotated formatted text
Follow this sample only if you want to import pre-annotated documents to tagtog when the input text was formatted when annotated.
This example shows how to send text to be formatted along with its annotations. The format used is formatted-plus-annjson. The input files are in Github, you can find a link below.
import requests
import sys
content_path = "files/formatted.txt"
annjson_path = "files/formatted.ann.json"
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"project": "yourProjectName", "owner": "yourUsername", "format": "formatted-plus-annjson", "output": "null"}
files=[("files", open(content_path)), ("files", open(annjson_path))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "formattedtext",
"filenames": ["formatted.ann.json", "formatted.txt"],
"names": ["formatted.ann.json", "formatted.txt"],
"rawInputSizeInBytes": 860,
"docid": "aAyUEVY5RCLzd8kdaOMg54fXXWj8-formatted",
"tagtogID": "aAyUEVY5RCLzd8kdaOMg54fXXWj8-formatted",
"result": "created",
"parsedTextSizeInBytes": 126
}],
"warnings": []
}
Examples: import pre-annotated PDF
This example shows how to import a PDF along with its annotations. The format used is default-plus-annjson as we want the PDF to use the default format and import annotations for this file. The input files are in Github, you can find a link below.
If you are rather annotating the PDF through tagtog’s internal plain.html (which contains the PDF’s text), take a look at this fully-working GitHub sample repository.
import requests
import sys
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"project": "yourProjectName", "owner": "yourUsername", "format": "default-plus-annjson", "output": "null"}
files=[("files", open('files/article.pdf', "rb")), ("files", open('files/article.ann.json'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "article.pdf",
"filenames": ["article.ann.json", "article.pdf"],
"names": ["article.ann.json", "article.pdf"],
"rawInputSizeInBytes": 1048119,
"docid": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"tagtogID": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"result": "created",
"parsedTextSizeInBytes": 83199
}],
"warnings": []
}
Examples: import a list of pre-annotated files
This example shows how to import a list of pre-annotated files. The format used is default-plus-annjson as we want each file to use the default format and to be pre-annotated by an annotation file.
The expected input are pair of content+ann.json files.
import requests
import sys
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"project": "yourProjectName", "owner": "yourUsername", "format": "default-plus-annjson", "output": "null"}
files=[("files", open('article.pdf', "rb")), ("files", open('article.ann.json')), ("files", open('item1.txt')), ("files", open('item1.ann.json'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
Response, output=null
{
"ok": 2,
"errors": 0,
"items": [{
"origid": "article.pdf",
"filenames": ["article.ann.json", "article.pdf"],
"names": ["article.ann.json", "article.pdf"],
"rawInputSizeInBytes": 1048119,
"docid": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"tagtogID": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"result": "created",
"parsedTextSizeInBytes": 83199
}, {
"origid": "item1.txt",
"filenames": ["item1.ann.json", "item1.txt"],
"names": ["item1.ann.json", "item1.txt"],
"rawInputSizeInBytes": 461,
"docid": "aGMgsSYn0VJlSHWgGD4zwsIvOqDG-item1.txt",
"tagtogID": "aGMgsSYn0VJlSHWgGD4zwsIvOqDG-item1.txt",
"result": "updated",
"parsedTextSizeInBytes": 128
}],
"warnings": []
}
Examples: import text pre-annotated by spaCy
This example shows how to generate a set of annotations with a spaCy model and send the pre-annotated text to tagtog. The model used is en_core_web_sm. We want to do NER and extract PEOPLE, ORG, and MONEY entities (see “Label Scheme”).
For more details, check out this step-by-step guide: Integrating tagtog and spaCy & the full GitHub repository.
import spacy
import json
import requests
import os
def get_class_id(label):
"""
Translates the spaCy label id into the tagtog entity type id
- label: spaCy label id
"""
choices = {'PERSON': 'e_1', 'ORG': 'e_2', 'MONEY': 'e_3'}
return choices.get(label, None)
def get_entities(spans, pipeline):
"""
Translates a tuple of named entity Span objects (https://spacy.io/api/span) into a
list of tagtog entities (https://docs.tagtog.com/anndoc.html#ann-json). Each entity is
defined by the entity type ID (classId), the part name where the annotation is (part),
the entity offsets and the confidence (annotation status, who created it and probabilty).
- spans: the named entities in the spaCy doc
- pipeline: trained pipeline name
"""
default_prob = 1
default_part_id = 's1v1'
default_state = 'pre-added'
tagtog_entities = []
for span in spans:
class_id = get_class_id(span.label_)
if class_id is not None:
tagtog_entities.append( {
'classId': class_id,
'part': default_part_id,
'offsets':[{'start': span.start_char, 'text': span.text}],
'confidence': {'state': default_state,'who': ['ml:' + pipeline],'prob': default_prob},
'fields':{},
# this is related to the kb_id (knowledge base ID) field from the Span spaCy object
'normalizations': {}} )
return tagtog_entities
MY_USERNAME = os.environ['MY_TAGTOG_USERNAME']
MY_PASSWORD = os.environ['MY_TAGTOG_PASSWORD']
MY_PROJECT = 'demo-spaCy'
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username=MY_USERNAME, password=MY_PASSWORD)
text = "Paypal Holdings Inc (PYPL) President and CEO Daniel Schulman Sold $2.7 million of Shares"
# Load the spaCy trained pipeline (https://spacy.io/models/en#en_core_web_sm) and apply it to text
pipeline = 'en_core_web_sm'
nlp = spacy.load(pipeline)
doc = nlp(text)
# Fill the ann.json
annjson = {}
annjson['anncomplete'] = False
annjson['metas'] = {}
annjson['relations'] = []
annjson['entities'] = get_entities(doc.ents, pipeline)
params = {'owner': MY_USERNAME, 'project': MY_PROJECT, 'output': 'null', 'format': 'default-plus-annjson'}
files=[('doc1.txt', text), ('doc1.ann.json', json.dumps(annjson))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
print(response.text)
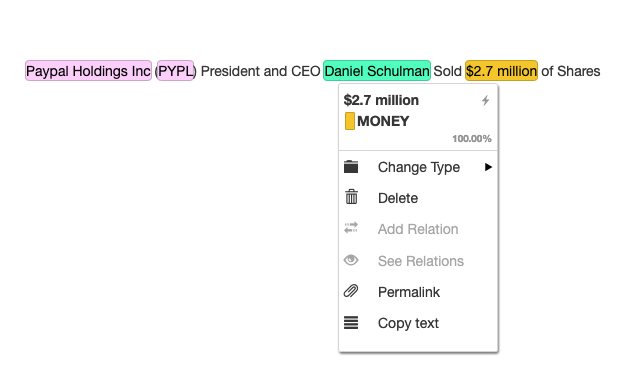
The resulting pre-annotated document visualized in tagtog editor
Replace annotations of existing document POST
You should use two files:
The original content file or the plain.html.
The annotations. You pass this as an ann.json.
If you use the original content file, it must have the same name as the original. If you want to use the plain.html, it should use the same name as the original plain.html.
Both files (content and annotations) should have the same name, except for the file extensions. For example: mydoc.txt and mydoc.ann.json. Or, if with a plain.html: mydoc-3243hdsfk3.plain.html and mydoc-3243hdsfk3.ann.json
If the original content doesn't exist in your project, the pre-annotated document will be also imported as new.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
files |
mydoc.txt, mydoc.ann.json | You need to upload in the same request both: the content file and the ann.json (annotations) files. | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use | |
output |
visualize |
null |
|
format |
anndoc |
anndoc |
Format of the pre-annotated document. List of supported pre-annotated formats: Pre-annotated formats |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
Examples: replace the annotations of an existing document using the original content
As you can see, this example is basically the same as the example to upload pre-annotated plain text. The only difference is that the original file should already exist in your project.
This example shows how to replace the annotations of an existing document (content + ann.json) to tagtog. If the original file doesn't exist in your project, it will be created.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null", 'format': 'default-plus-annjson'}
files = [("files", open('/annotated-docs/mydoc.txt')), ("files", open('/annotated-docs/mydoc.ann.json'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "mydoc.txt",
"filenames": ["mydoc.ann.json", "mydoc.txt"],
"names": ["mydoc.ann.json", "mydoc.txt"],
"rawInputSizeInBytes": 1048119,
"docid": "aqXHSykmx2gmA9AJXW38OAl0DnTe-mydoc.txt",
"tagtogID": "aqXHSykmx2gmA9AJXW38OAl0DnTe-mydoc.txt",
"result": "updated",
"parsedTextSizeInBytes": 83199
}],
"warnings": []
}
Examples: replace the annotations of an existing document using plain.html
If it is more convenient, you can use the plain.html version of the original file (plain text representation of the file) to replace the annotations on the original file.
This example shows how to replace the annotations of an existing document (plain.html + ann.json) to tagtog. Please notice that the original file should already exist in your project. tagtog will automatically identify the original file and replace its annotations.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "output": "null", 'format': 'anndoc'}
files=[("files", open('files/article.html')), ("files", open('files/article.ann.json'))]
response = requests.post(tagtogAPIUrl, params=params, auth=auth, files=files)
Response, output=null
{
"ok": 1,
"errors": 0,
"items": [{
"origid": "article.pdf",
"filenames": ["article2.ann.json", "article2.html"],
"names": ["article2.ann.json", "article2.html"],
"rawInputSizeInBytes": 86729,
"docid": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"tagtogID": "aqXHSykmx2gmA9AJXW38OAl0DnTe-article.pdf",
"result": "updated",
"parsedTextSizeInBytes": 83199
}],
"warnings": []
}
Search documents in a project GET
You can search using the documents API. Search across your project and retrieves the matching documents. You can use it to augment your own search engine or simply create a new one. It is also very simple to use the search API to display statistics.
Learn how to build search queries here.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
search |
entity:GGP:P02649 or folder:pool | Search query. Learn how to build queries here. | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
page |
0 | 1 | Number: page number in a paginated search. |
output |
search |
search |
You can choose between
|
Search response format
Response format for search queries.
{
"version": "String: this format's version, e.g. 0.7.0",
"search": "String: user search query",
"totalFound": "Number: total number of documents that match the search query",
"pages": {
//the search is paginated
"current": "Number: paginated search's current page number (0-indexed)",
"previous": "Number: paginated search's previous page; -1 if current page == 0",
"next": "Number: paginated search's current page; -1 if current page is the last page",
"numPages": "Number: number of pages; at least 1",
"pageSize": "Number: number of document elements in the page; always 50"
}
"docs":
[
{
"id": "String: full tagtogID -- Use this to download the document",
"filename": "String: filename of originally uploaded file",
"header": "String: title if the document has a natural title or otherwise an excerpt of the text's start",
"created": "String: date for the document' upload time, in ISO_INSTANT format, e.g. 2021-07-15T16:21:25.750Z",
"updated": "String: date for the document' last update, in ISO_INSTANT format, e.g. 2021-07-16T16:28:17.285Z",
"anncomplete": "Boolean: status for the document's annotation completion",
"members_anncomplete": ["String Array: members' usernames who completed (confirmed) their annotations"],
"members_assigned": ["String Array: members' usernames who were asssigned to this document"],
"folder": "String: folder path where the document is located; e.g. `pool/mySubFolder`"
},
//next documents in the array of results...
]
}
Examples: search using search queries
This example searches across all your folders to find documents that have at least one entity normalized to the gene P02649.
curl -u yourUsername:yourPassword 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&search=entity:GGP:P02649'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", "search": "entity:GGP:P02649"}
response = requests.get(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&search=entity:GGP:P02649', {
method: 'GET',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword')},
}).then(response => response.text()).then(text => {
console.log(text);
}).catch(function(error) {
console.log('Error: ', error);
});
search response
{
"version": "0.7.0",
"search": "entity:GGP:P02649",
"totalFound": 1,
"pages": {
"current": 0,
"next": -1,
"numPages": 1,
"pageSize": 50,
"previous": -1
},
"docs": [
{
"anncomplete": false,
"created": "2021-07-15T16:34:41.720Z",
"filename": "text.txt",
"folder": "pool",
"header": "tagtog is awesome ;-)",
"id": "aE7DVwx5pj.KxZtSadEce0HkNGk0-text.txt",
"members_anncomplete": [
"user-A"
],
"members_assigned": [
"user-C",
"user-A"
],
"updated": "2021-07-15T16:35:34.428Z"
}
]
}
csv response
docid,anncomplete
abPz9JKO2jdP9XKbn4Beuh3rk3Y4-text,false
aPvgZql3RogPu90jVkoV7rZODU8u-text,false
ap_FCtCdahae2jMD_opHUD9f7lM8-text,true
aMHKzF_lIoNrdh9pAx298njgIezy-text,false
Get existing documents GET
You can use the API to export documents. You need the id of the document to get it. If you don't have this id, you can find it using the search feature. You can export only 1 document within each request.
Specify the output parameter to define the output format (e.g. ann.json, html)
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
output |
visualization |
ann.json |
The format of the output you want to be returned by the API. API output formats. |
idType |
tagtogID |
tagtogID |
Type of Id. List of idTypes |
ids |
aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text | The id of the document you want to download. Note, the parameter is called "ids" for historical reasons. In the future, we might also allow to download multiple files at once. | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use |
Optional Parameters
| Name | Default | Example | Description |
|---|---|---|---|
member |
master |
John | Annotation version, either |
Examples: get the annotations of a document by document id
This example retrieves the annotations of a document in ann.json format. As the member parameter is not defined, the master version of the annotations is served. Notice that we don't use the parameter idType because it defaults to tagtogID, the type of the id used.
curl -u yourUsername:yourPassword 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&ids=aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text&output=ann.json'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'ids':'aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text', "output": "ann.json"}
response = requests.get(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&ids=aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text&output=ann.json', {
method: 'GET',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword')},
}).then(response => response.text()).then(text => {
console.log(text);
}).catch(function(error) {
console.log('Error: ', error);
});
Response, output=ann.json
{
"anncomplete":false,
"sources":[{"name":"PMID","id":"23596191","url":"http://www.ncbi.nlm.nih.gov/pubmed/23596191"}],
"entities":
[
{"classId":"e_1","part":"s1h1","offsets":[{"start":60,"text":"RETICULATA-RELATED"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":315,"text":"RETICULATA-RELATED"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":335,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":444,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":561,"text":"PROTEIN"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3289},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q8IVL6","url":null},"recName":"Prolyl 3-hydroxylase 3","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.3289}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1127,"text":"rer1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4836},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4836}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1265,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1303,"text":"RER1"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"O15258","url":null},"recName":"Protein RER1","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.7737}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1391,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1587,"text":"RER"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q13123","url":null},"recName":"Protein Red","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.6519}}}},
{"classId":"e_1","part":"s2p1","offsets":[{"start":1591,"text":"proteins"}],"confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4073},"fields":{},"normalizations":{"n_2":{"source":{"name":"SwissProt","id":"Q15517","url":null},"recName":"Corneodesmosin","confidence":{"state":"pre-added","who":["ml:dpeker"],"prob":0.4073}}}}
],
"metas":{},
"relations":[],
"annotatable":{"parts":["s1h1","s2h1","s2p1"]}
}
Examples: get the member's annotations of a document by document id
This example retrieves the annotations of tagtog document in ann.json format. A document can have different annotation versions, in this case we want the version of the annotations from the member John
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'ids':'aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text', 'member': 'John', "output": "ann.json"}
response = requests.get(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
Examples: get the original document by document id
This example download the original document (format orig) given a document id. Notice that we don't use the parameter idType because it defaults to tagtogID, the type of the id used.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'ids':'aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text', "output": "orig"}
response = requests.get(tagtogAPIUrl, params=params, auth=auth)
if response.status_code == 200:
with open("mydoc.pdf", "wb") as f:
f.write(responseGet.content)
Examples: get the html version of a document by document id
This example download the HTML version of a document (format html) given a document id. The HTML follows the plain.html specification, which is the text representation of the original document, used to calculate the offsets of the annotations.
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
docId = "aVTjgPL0x5m_xgJr3qcpfXcSoY_q-text"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'ids':docId, "output": "html"}
response = requests.get(tagtogAPIUrl, params=params, auth=auth)
if response.status_code == 200:
with open(docId + '.html', 'wb') as f:
f.write(responseGet.content)
Delete documents DELETE
Delete documents by search
You can delete documents in your project using the API. Fine-tune the search parameter to delete only those documents returned by the search query.
This request returns the number of documents deleted.
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
search |
entity:GGP | Search query to list the documents to remove. Learn how to build queries here | |
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use |
Examples: delete documents using a search query
This example deletes all documents that contain at least one entity of type gene.
curl -u yourUsername:yourPassword -X DELETE 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&search=entity:gene'
import requests
tagtogAPIUrl = "https://www.tagtog.com/-api/documents/v1"
auth = requests.auth.HTTPBasicAuth(username="yourUsername", password="yourPassword")
params = {"owner": "yourUsername", "project": "yourProjectName", 'search':'entity:gene'}
response = requests.delete(tagtogAPIUrl, params=params, auth=auth)
print(response.text)
fetch('https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&search=entity:gene', {
method: 'DELETE',
headers: {'Authorization' : "Basic " + btoa('yourUsername' + ":" + 'yourPassword')},
}).then(response => response.text()).then(text => {
console.log(text);
}).catch(function(error) {
console.log('Error: ', error);
});
Response
4 //number of documents deleted
Delete document by id
Delete a single document given its tagtog document id.
This request returns the number of documents deleted (1 if the document was successfully deleted, 0 otherwise).
Input Parameters
| Name | Default | Example | Description |
|---|---|---|---|
idType |
tagtogID (mandatory) |
||
ids |
aEVD52vVm.s2zdTmzK_ACNqH7Z1u-text | ||
project |
yourProjectName | Name of the project | |
owner |
yourUsername (in this example we assume the user is also the owner of the project) | Owner of the project you want to use |
Examples: delete a document by document id
This example deletes a document given by its (tagtog) document id.
curl -u yourUsername:yourPassword -X DELETE 'https://www.tagtog.com/-api/documents/v1?owner=yourUsername&project=yourProjectName&idType=tagtogID&ids=yourDocumentTagtogID'
output parameter
idType parameter
Possible values for the parameter are described below.
| Name | Description |
|---|---|
tagtogID |
This is the default value. tagtog-internal document id or docid (e.g. ai1AzDk4wQzbL.BKzlrA_CrK8gJi-text). Its use implicitly means that the document already exists in the associated project. |
PMID |
PubMed ID. |
PMCID |
PubMed Central ID. |
Manage annotation versions
🤠𝛂 These APIs are in alpha, and can change at any moment. We give early access for your benefit.
Merge the annotations of a document (Automatic Adjudication)
Merge the confirmed members’ annotations of a document.
This assumes that the document was confirmed by at least one member. If the given document was no confirmed by any member yet, the response will return an error.
You can know which documents have at least one version confirmed using the search API.
- Method:
POST - Endpoint:
/-api/documents/versions/v0/merge?owner=...&project=...
Input (parameters)
Body: None
| Type | Name | Default | Example | Description |
|---|---|---|---|---|
| Query | docid |
“xxx-text.txt” | tagtog’s id of the document to merge. | |
| Query | strategy |
“union_v1” | Merging strategy, in: union_v1, intersection_v1, majority_v1, best_iaa_v1` |
|
| Query | saveTo (OPTIONAL) |
N.A. | “master” | project member’s username (incl. “master”) to save the annotation merging result to. The merging result is always returned in the body response as an ann.json object. Additionally, if you set this parameter, the result annotations will be saved in the given member. |
Output
Successful status code: 200 (OK)
Payload: JSON (application/json)
The merging’s ann.json result.
Copy the annotations from a member to another one
This assumes that the member to copy the annotations from (the source) actually has some annotations. It’s also possible to copy the annotations of master to another member or viceversa.
NOTE: the copying of annotations is always disallowed if the member to save to (the target) already had confirmed annotations.
NOTE: Even if the source annotations were confirmed, the resulting annotations in target will always be unconfirmed.
- Method:
POST - Endpoint:
/-api/documents/versions/v0/copy?owner=...&project=...
Input (parameters)
Body: None
| Type | Name | Default | Example | Description |
|---|---|---|---|---|
| Query | docid |
“xxx-text.txt” | tagtog’s id of the document (annotations) to copy. | |
| Query | from |
“user-A” | source; project member’s username (incl. “master”) to copy the annotations from. | |
| Query | saveTo |
“user-B” | target; project member’s username (incl. “master”) to save the annotations to. The resulting annotations are always returned in the body response as an ann.json object. |
Output
Successful status code: 200 (OK)
Payload: JSON (application/json)
The final ann.json now stored in the saveTo member. This ann.json is always unconfirmed, nevermind the status of the source annotations.
API Clients
Python tagtog script
If you want to use an already built API client. You have the ![]() tagtog python API script to do many common operations in tagtog using the API: upload (also folders), search, delete, and download documents!
tagtog python API script to do many common operations in tagtog using the API: upload (also folders), search, delete, and download documents!
usage: tagtog [-h] {upload,search,download,delete} ...
tagtog official script to Upload & Search & Download & Delete documents.
Version: 0.3.0
Author: tagtog Sp. z o.o.
Website: https://www.tagtog.com
API documentation: https://docs.tagtog.com/API_documents_v1.html
positional arguments:
{upload,search,download,delete}
upload Upload files to tagtog
search Search documents by query, e.g. `*` (all)
download Download documents by search query, e.g. `updated:[NOW-1DAY to NOW]
delete Delete documents that match a search query, e.g. `docid:aZ8wXRHvqyw7tjBQW8NXMTPQ0S.C-test.md` (to delete a specific doc)
optional arguments:
-h, --help show this help message and exit
Search
It uses the API to search documents in your project. Parameters can be consulted here or using tagtog.py search --help. You can learn how to build search queries here
The example below retrieves all the documents from your project.
python3 tagtog.py search "*" -u yourUsername -w yourPassword -o yourUsername -p yourProjectName
Upload
It uses the API to upload documents to your project.
Upload PMIDs
Parameters can be consulted here or using tagtog.py upload --help.
The example below upload the abstract from two PMIDs to your project. Remember to indicate which is the type of id (--idType or -i) for the document.
python3 tagtog.py upload 29539636,29531059 -u yourUsername -w yourPassword -o yourUsername -p yourProjectName -i PMID
Upload files
Parameters can be consulted using tagtog.py upload --help. You must include the parameter --extension or -e to indicate the extension of the files to upload (e.g. txt, pdf, etc.). These are the input files supported
The example below upload the PDF documents of a folder, to your project.
python3 tagtog.py upload ./myfolder -u yourUsername -w yourPassword -o yourUsername -p yourProjectName --extension pdf
The example below upload a single file to your project.
python3 tagtog.py upload ./myfile.txt -u yourUsername -w yourPassword -o yourUsername -p yourProjectName
Download
It uses the API to download documents from your project. In one call to the script you can download all documents matching a search query.
Parameters can be consulted using tagtog.py download --help.
You can indicate the folder where you can store the downloaded documents using the parameter --output_folder (it defaults to the folder where the script is running).
Use the parameter --output or -t to indicate the output type for the downloaded files.
You can learn how to build search queries here
The example below download the annotations (ann.json) for all the documents in a project.
python3 tagtog.py download "*" -u yourUsername -w yourPassword -o yourUsername -p yourProjectName --output_folder ./myDownloadFolder -t ann.json
